Visualize NDVI in Microsoft Planetary Computer

What is Normalized Difference Vegetation Index (NDVI)?
The Normalized Difference Vegetation Index (NDVI) is quantify the vegetation by measuring the difference between near-infrared and red bands.
NDVI = (NIR-Red)/(NIR+Red)
NDVI value ranges between 1 to +1. The positive value tends to dense vegetation and negative value represents non-vegetation like water-bodies, snow etc.
In this tutorial, I will show you how to create an NDVI map from Landsat data in Microsoft Planetary Computer.
Step – 1: Environment setup
import pystac_client
import planetary_computer
import odc.stac
import matplotlib.pyplot as plt
from pystac.extensions.eo import EOExtension as eoStep – 2: Access to Planetary Computer Data
catalog = pystac_client.Client.open(
"https://planetarycomputer.microsoft.com/api/stac/v1",
modifier=planetary_computer.sign_inplace,
)Step – 3: Choose an area and time of interest
bbox_of_interest = [89.419391,22.732522,89.671905,22.853430] # Bounding box of study
time_of_interest = "2022-01-01/2022-12-31" # Define start and end dateStep – 4: Define search parameters
search = catalog.search(
collections=["landsat-c2-l2"],
bbox=bbox_of_interest,
datetime=time_of_interest,
query={"eo:cloud_cover": {"lt": 1}},
)
Here, collections indicates the dataset, query to define cloud cover of image. The “lt” means less than. We set the cloud cover less than 1% means it will return the image collection where each image have less than 1% cloudy.
Step – 5: Identify the length of image collection
items = search.item_collection()
print(f"Returned {len(items)} Items")
Returned 25 Items
selected_item = items[5] # min(items, key=lambda item: eo.ext(item).cloud_cover)
print(
f"Choosing {selected_item.id} from {selected_item.datetime.date()}"
+ f" with {selected_item.properties['eo:cloud_cover']}% cloud cover"
)We randomly select the image which index number is 5. The print options shows the image id, date of acquisition and its cloud cover.
Choosing LC09_L2SP_138044_20221114_02_T1 from 2022-11-14 with 0.05% cloud cover
Step – 6: Exploring image bands
max_key_length = len(max(selected_item.assets, key=len))
for key, asset in selected_item.assets.items():
print(f"{key.rjust(max_key_length)}: {asset.title}")
qa: Surface Temperature Quality Assessment Band
ang: Angle Coefficients File
red: Red Band
blue: Blue Band
drad: Downwelled Radiance Band
emis: Emissivity Band
emsd: Emissivity Standard Deviation Band
trad: Thermal Radiance Band
urad: Upwelled Radiance Band
atran: Atmospheric Transmittance Band
cdist: Cloud Distance Band
green: Green Band
nir08: Near Infrared Band 0.8
lwir11: Surface Temperature Band
swir16: Short-wave Infrared Band 1.6
swir22: Short-wave Infrared Band 2.2
coastal: Coastal/Aerosol Band
mtl.txt: Product Metadata File (txt)
mtl.xml: Product Metadata File (xml)
mtl.json: Product Metadata File (json)
qa_pixel: Pixel Quality Assessment Band
qa_radsat: Radiometric Saturation and Terrain Occlusion Quality Assessment Band
qa_aerosol: Aerosol Quality Assessment Band
tilejson: TileJSON with default rendering
rendered_preview: Rendered preview
Step - 7: Band selection
bands_of_interest = ["nir08", "red", "green", "blue", "qa_pixel"]
data = odc.stac.stac_load(
[selected_item], bands=bands_of_interest, bbox=bbox_of_interest
).isel(time=0)
data
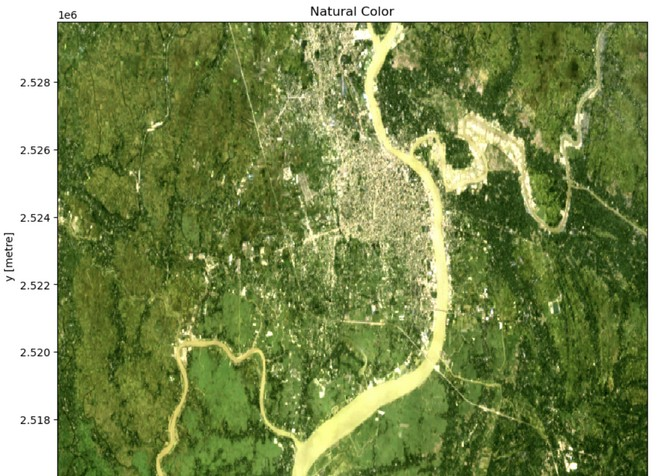
Step - 8: Plot and visualize the raw image
fig, ax = plt.subplots(figsize=(10, 10)) // Define plot size
data[["red", "green", "blue"]].to_array().plot.imshow(robust=True, ax=ax)
ax.set_title("Natural Color");
Step – 9: Calculate and visualize NDVI
red = data["red"].astype("float")
nir = data["nir08"].astype("float")
ndvi = (nir - red) / (nir + red)
fig, ax = plt.subplots(figsize=(10, 6))
ndvi.plot.imshow(ax=ax, cmap="viridis")
ax.set_title("NDVI");
GitHub Source Code Link - NDVI Planetary Computer
If you like this tutorial, please share with your connections  .
.
Share To
About Author

- Kamal Hosen
Geospatial Developer | Data Science | PythonA passionate geospatial developer and analyst whose core interest is developing geospatial products/services to support the decision-making process in climate change and disaster risk reduction, spatial planning process, natural resources management, and land management sectors. I love learning and working with open source technologies like Python, Django, LeafletJS, PostGIS, GeoServer, and Google Earth Engine.